Illustration (click to hide):
Project Description
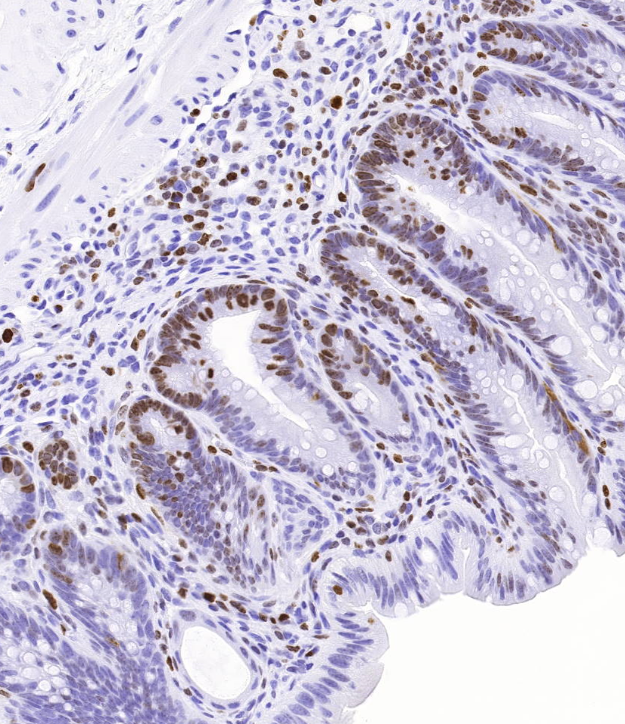
Colorectal cancer is the third most common cancer in both men and women in Sweden and the third most common cause of cancer related death. Even though a number of genetic alterations have been linked to colorectal cancer we still do not know the full complement of molecular lesions that are individually necessary – and together sufficient – to cause the disease. In a recent genetic characterization of a large set of colorectal cancer, we identified a potential colorectal cancer driver event. Using mouse models we are now exploring its role both during normal intestinal homeostasis and intestinal tumorigenesis. The effect of the genetic alteration on the normal stem cell population and the continuous renewal of the intestinal epithelium is examined in several different ways, for example by counting and comparing the number of proliferating cells per crypt in intestines with and without the genetic alteration. The Ki67-positive proliferating cells are counted using a script in Fiji that can differentiate brown DAB-stained proliferating cells from blue hematoxylin counterstained nuclei. We are also exploring the genetic alteration’s potential involvement in colorectal cancer by examining its impact on tumor formation in the ApcMin/+ intestinal tumor model. The number and sizes of intestinal polyps/tumors formed are scored and compared to that of control ApcMin/+ mice. Polyp size measurements are done in Qupath. Gisele Miranda at the SciLifeLab BioImage Informatics Facility helped with optimizing the polyp measurement method and designed the script in Fiji we use to count DAB-stained cells.
Project Information
-
BIIF Principal Investigators
- Gisele Miranda
External Authors
Lauri Aaltonen, Åsa Kolterud -
Date
2019-09-05 🠚 Current