Illustration (click to hide):
Project Description
In this project, we set out to establish a method to detect multiple protein interactions and protein activation states in different types of cancerous tissue. Building on our long tradition of molecular tools development, we will establish an in situ proximity ligation assay (isPLA) technology to assess protein status in clinical samples. Image analysis represents a central aspect of this project, and we wish to acquire signal characteristics directly from data and relate results to other image parameters such as tissue morphology and other molecular characteristics. Our aim is to develop an in situ PLA method that will generate rolling circle products labelled with fluorophores upon affinity binding to each protein of interest, generating multiple signals in each tissue sample. These multiple fluorophore signals will be detected using an imaging system that translates the signals directly from the image data to localized signals to explore spatial patterns in each tissue.
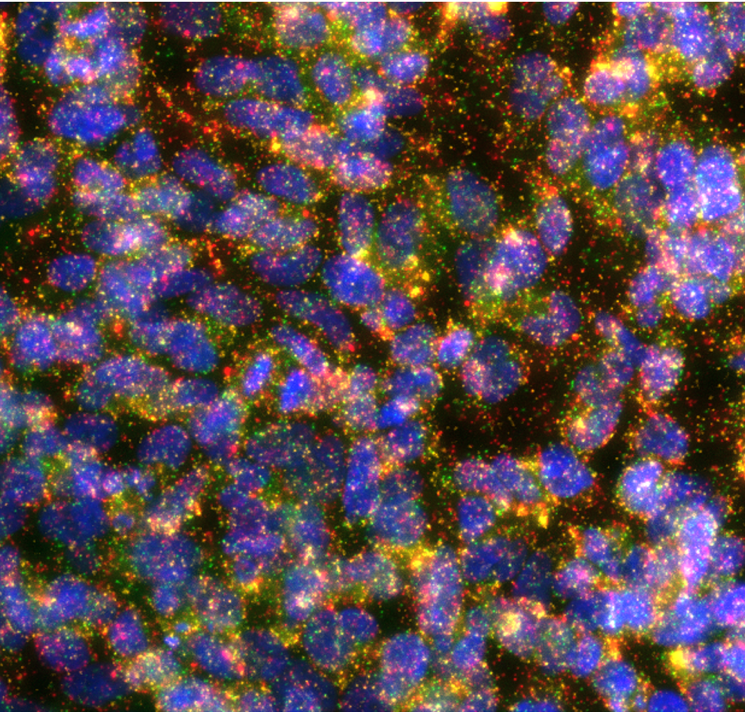
Currently, we are validating the methods using different breast cancer cell lines. And we will need the help from BioImage Informatics Facility for developing a pipeline for analyzing the images (Figure 1 as an example) taking from the multiplexed isPLA assays.
(Figure: Interactions of ERBB family receptors in SKBR3 cells. EGFR-HER2: green, HER3-HER2: yellow, HER4-HER2: red, nuclei: blue.)
Project Information
-
BIIF Principal Investigators
- Christophe Avenel
External Authors
Bo Xu -
Date
2021-06-01 🠚 Current